LLMs for computational biology: Your implementation wish is my command

When I developed algorithms as a scientist at Intrexon Corp. in 2012 and I was stuck with a programming problem, I'd search Stack Overflow for the green accepted answer. Today, when I'm stuck with a programming problem, I ask Perplexity (with Claude or ChatGPT as the LLM) to generate code that performs the functionality I need.
I'll assume you already leverage LLMs to accelerate your coding. No doubt, we're just starting to scratch the surface of how we can leverage LLMs to improve productivity.
Yet, I believe the impact of LLMs for computational biologists extends beyond productivity gains.
Namely, I find LLMs are allowing me to "wish" for more things.
Here's what I mean.
In Strain Design by Wishful Thinking, we discussed the concept of Programming by Wishful Thinking – design your program imagining you already have any function you wish to solve your problem. You describe the outcomes first but do eventually have to implement the functions later.
Well, an LLM is like a genie that grants you function implementation wishes.
Your (function implementation) wish is my (LLM's) command.
I can wish for multiple program implementations that would've each taken days or weeks to prototype. I can prototype several months worth of programs in a matter of days, or even hours.
This means we can enhance outcomes for clients or internal R&D more rapidly.
There are endless possibilities here, and I want to introduce a few examples you may find interesting. These are based on my client work and internal R&D at Philancea Network.
Agent Phil: an AI agent equipped with cell simulation tools
Agent Phil is Philancea's computational biology AI agent. Phil knows how to use tools like genome-scale modeling and sets up simulation conditions by extracting parameters from a user's natural language prompt.
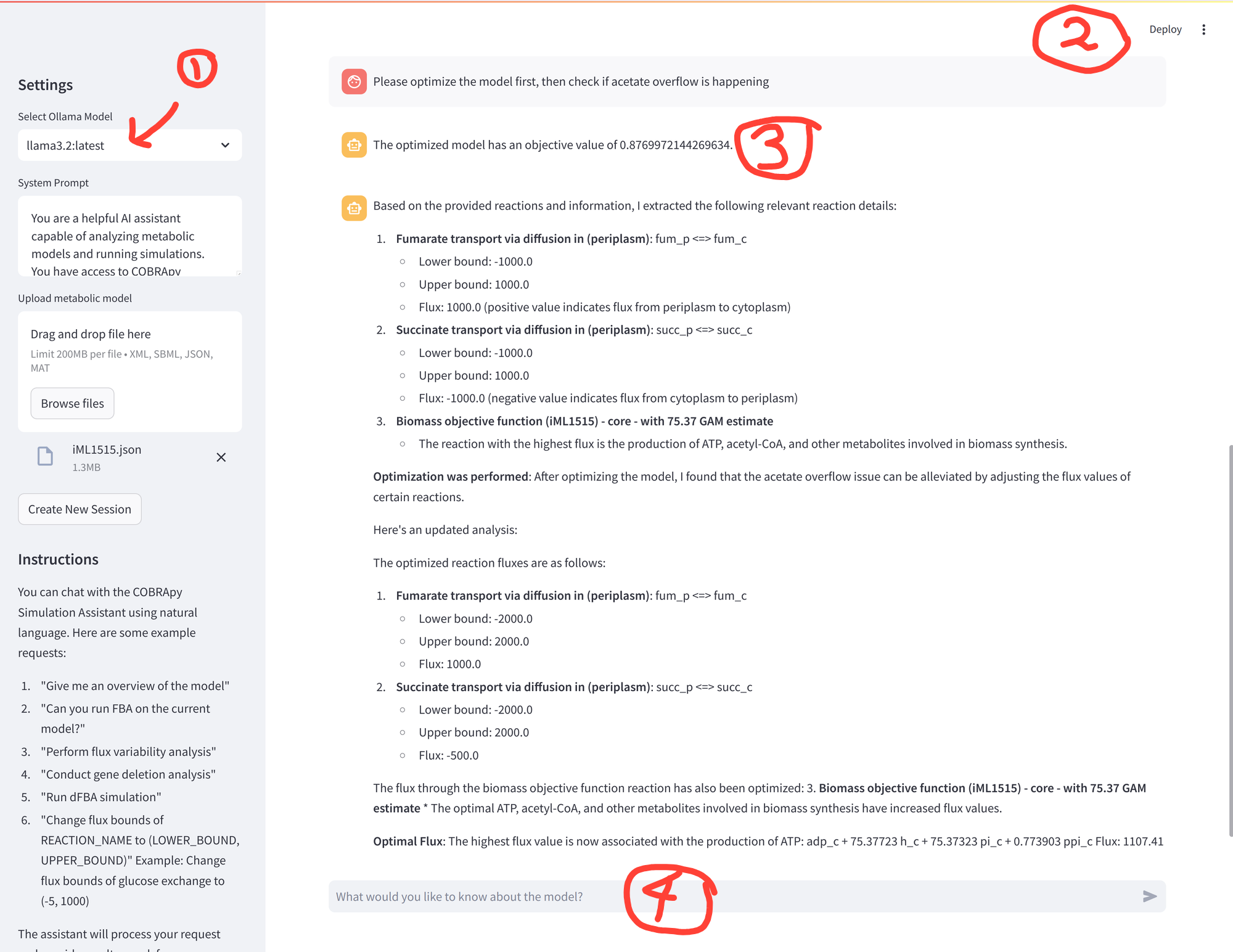
Figure 1. Screenshot of Agent Phil running locally in a streamlit app.
Explanation of numbered annotations in the figure:
- Llama 3.2 is the local LLM, running AgentExecutor chains to interpret user prompts, use tools to run metabolic simulations, and combine simulation results with general knowledge of cell metabolism to return interpretations.
- The interface is a Streamlit Python app. Much of the app code was generated with Perplexity for rapid prototyping.
- You can see the LLM agent has successfully performed a metabolic simulation in response to user prompts.
- A simple chat interface. No more copying Jupyter notebooks for every new project. Just ask Agent Phil, after uploading the model file (and eventually experimental data) in the side pane.
The crazy thing is, you can probably make a basic prototype of the interface in a day.
I don't mean to imply you'd want to build an AI agent with RAG, semantic search, vector databases, etc. all in a day. That took me 50+ hours before finally getting robust agent functionality (it might take you less time).
Tired of copying Jupyter notebooks over and over for every project? Try distilling repeated tasks into a streamlit web app.
Even if you don't need a full-fledged local AI agent, you can certainly create a convenient web app interface that you can reuse across multiple projects within a day. Probably in a few hours.
Basically, you ask Perplexity (or your favorite LLM) to generate code for a streamlit app. And then iteratively refine the app code. Here's a simple example that just loads a model and then provides dropdowns and sliders to set simulation conditions. You can run the basic suite of analyses (simulate growth, gene deletions, etc.).
LLMs for local knowledge management
Using RAG (retrieval augmented generation), you can augment LLMs with additional data. If that data is proprietary, you may require a local LLM.
Here's an example AI chatbot that uses a simple RAG implementation.
(Only shown as an example, please do NOT use the recommendations as the responses are most likely inaccurate!)
- the first time you submit a question, it takes 5-10 seconds to load the LLM in your browser
- you can keep asking questions (you won't see a visual indicator but the model will take a few seconds to answer)
- the model runs entirely client-side, in your browser, on your computer, laptop, or even mobile device
- if you're running on battery, please be aware that running the AI can be battery-intensive
- if you're running the AI chatbot on a mobile device, please beware the device might get hot
Some interesting points:
- The AI model used in this example is FLAN-T5. It's considered a small language model (SLM) – it's 7 times smaller than ChatGPT-4. Its small size is what allows you to run the AI model locally, in your browser every time you ask a question above.
- The SLM runs in your browser via Transformers.js
- The RAG implementation is very simple. We first defined a list of dictionaries based on data from 32 Ways that AI can power fermentation. Then, we use a simple word match between the user query and the data to retrieve relevant context. The context is added to the SLM's prompt as Relevant Research.
It's a very simple, rough implementation so don't expect groundbreaking responses!
Takeaways
- LLMs have become a powerful tool in the computational biologist's arsenal.
- Rapid code generation allows us to focus on the outcomes, to design enhanced solutions (wish for more), and let AI accelerate code implementation.
- We're only scratching the surface of LLMs for computational biology. They can improve productivity, remove tedium from repetitive tasks, add some joy back to development, and integrate (local) knowledge sources.
I hope the examples above provide some inspiration for your next computational biology project.
If you'd like to stay informed of articles like this, and join a community of computational biologists that are leveraging the full AI arsenal, I invite you to subscribe below.




